Publication Date: January 22, 2025
Authors:
Mohammad Ghazi Vakili, Christoph Gorgulla, Jamie Snider, AkshatKumar Nigam, Dmitry Bezrukov, Daniel Varoli, Alex Aliper, Daniil Polykovsky, Krishna M. Padmanabha Das, Huel Cox III, Anna Lyakisheva, Ardalan Hosseini Mansob, Zhong Yao, Lela Bitar, Danielle Tahoulas, Dora Čerina, Eugene Radchenko, Xiao Ding, Jinxin Liu, Fanye Meng, Feng Ren, Yudong Cao, Igor Stagljar, Alán Aspuru-Guzik & Alex Zhavoronkov
Abstract:
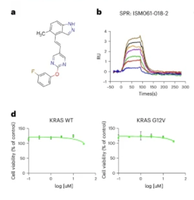
We introduce a quantum–classical generative model for small-molecule design, specifically targeting KRAS inhibitors for cancer therapy. We apply the method to design, select and synthesize 15 proposed molecules that could notably engage with KRAS for cancer therapy, with two holding promise for future development as inhibitors. This work showcases the potential of quantum computing to generate experimentally validated hits that compare favorably against classical models.
Related links: